“Over the decade I have used this technology, I have saved the lives of patis with unknown fungal infections”
Caroline Fife, MD, FAAFP, CWS
Medical Director, Wound Clinic at St. Luke’s Health, The Woodlands Hospital
Watch clinician testimonials
Diagnosing Challenging Wound Infections
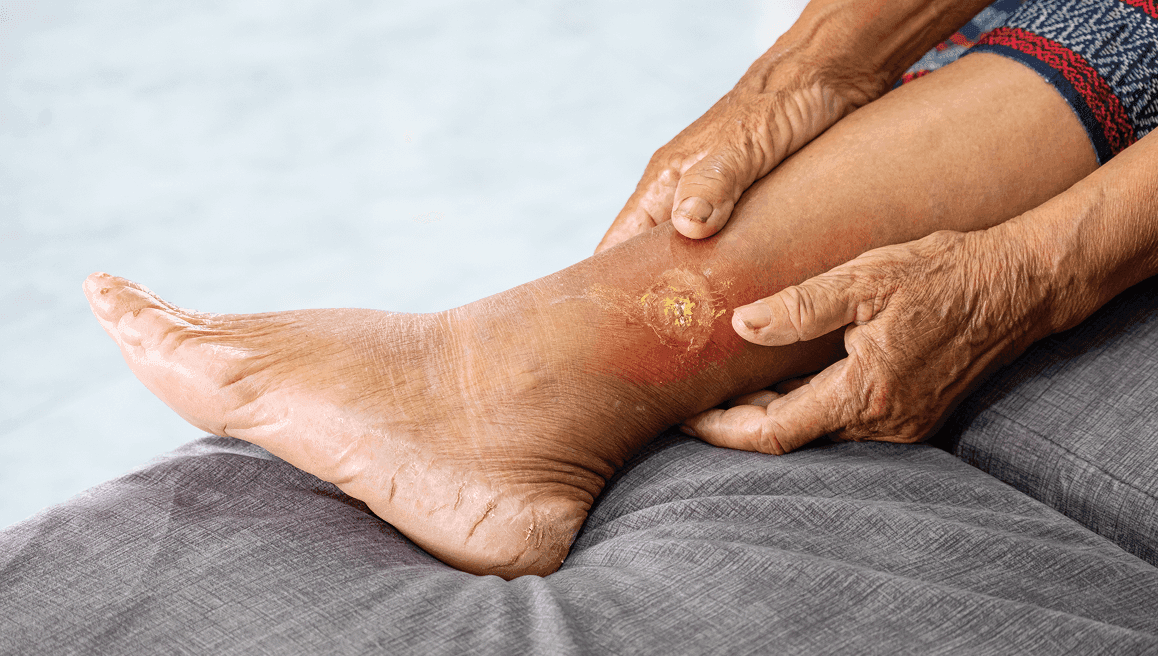
Chronic wound infections such as diabetic foot ulcers, pressure injuries, venous leg ulcers, and non-healing surgical wounds pose significant clinical challenges due to their frequent polymicrobial nature and the presence of biofilms. Biofilms protect microbial communities from immune responses and antimicrobial therapies, contributing to persistent infections and resistance. These wounds often involve mixed bacterial and fungal pathogens that are difficult to detect with standard culture methods, which frequently miss anaerobes, fungi, or microbes embedded in biofilms. Delayed or inadequate treatment can lead to serious complications, including systemic infection, limb loss, or prolonged hospitalization.
Our Solution:
MicroGenDX provides rapid microbial DNA sequencing to improve wound infection diagnostics and support better clinical outcomes. Our WoundKEY test is validated for swabs, tissue, and fluid samples to support the diagnosis and management of:
- Chronic wounds
- Diabetic foot ulcers
- Osteomyelitis
- Pressure injuries
- And more
Powered by next-generation sequencing (NGS) and qPCR, WoundKEY enables high-sensitivity detection of bacterial and fungal pathogens even in polymicrobial, biofilm-associated, or culture-negative infections. Resistance gene profiling supports earlier, more targeted antimicrobial therapy, helping clinicians implement effective, personalized wound management strategies and reduce the risk of delayed healing or complications.
Clinical Insights From our Wound Samples
Extensive Real-World Experience:
A study sequenced 2,963 chronic wound samples (DFUs, VLUs, pressure and surgical wounds) one of the largest wound-microbiome datasets¹.
Broad Microbial Diversity Identified:
Across those wounds, Staphylococcus spp. dominated 63 % of cases, Pseudomonas spp. 25 %, alongside a high prevalence of strict anaerobes and commensals that cultures routinely miss¹.
High Prevalence of Polymicrobial Infections:
Most chronic wounds are polymicrobial; in diabetic-foot osteomyelitis, NGS detected anaerobes in 87 % of bone samples versus 23 % by culture, showing how often standard methods under-report key pathogens².
Detection of Fastidious or VNBC (Viable but Non-Culturable) Pathogens:
NGS routinely uncovers hard-to-culture bacteria, biofilm constituents, and rare fungi e.g., a case of Rhizopus mucormycosis³ invisible to culture but caught by MicroGenDX, enabling life-saving therapy.
Recommended use of Next-Generation DNA Sequencing in Wound care

Global Wound Biofilm Expert Panel
Consensus guidelines from a Global Wound Biofilm Expert Panel include the use of DNA diagnostics in the first 1-4 days of chronic wound therapy as part of early intervention.

International wound infection institute
“DNA sequencing techniques can more precisely identify microbial species in a wound specimen, including microbes not identified by culture-based techniques… Standard clinical microbiology laboratory results may only provide information about a small subset of the total bacterial species that are present, particularly in chronic wounds. ” *

German S1 Guideline 2023
Advise to examine samples by microscopy, culture and/or molecular testing, adding that PCR or DNA sequencing to allows species-level identification of the pathogen. **
- * International Wound Infection Institute. (2022). Wound infection in clinical practice: Principles of best practice (International consensus update, 3rd ed.). Wounds International.
- ** Nenoff P, Reinel D, Mayser P et al. S1 guideline onychomycosis. J Dtsch Dermatol Ges (2023)
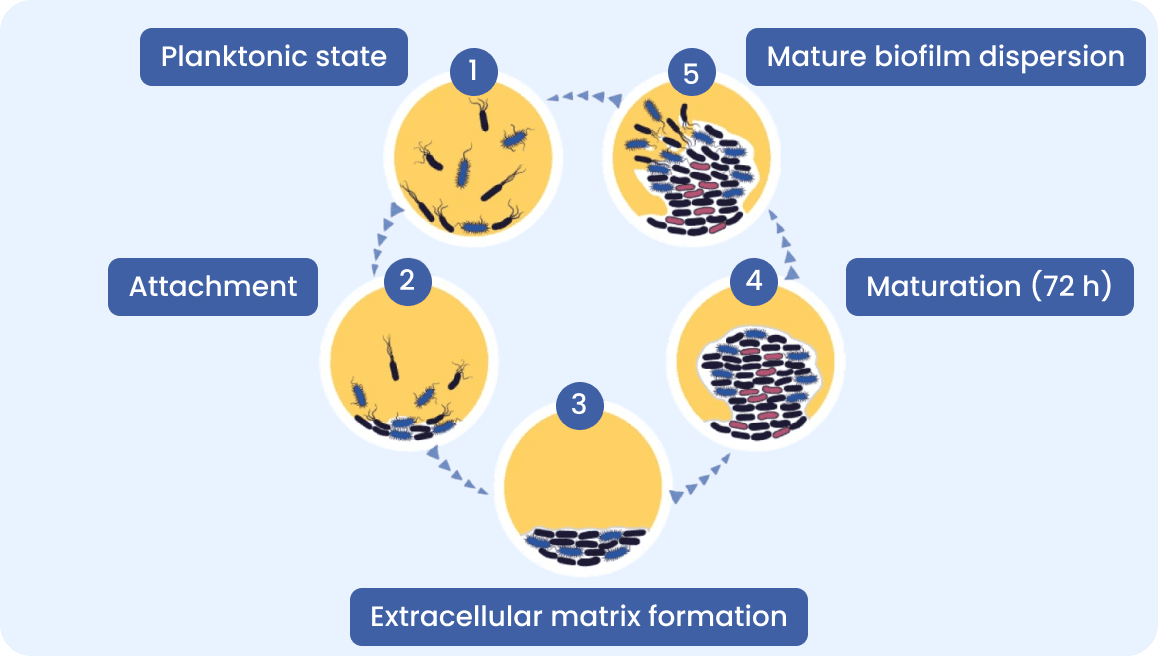
Biofilm Role in Chronic Wound infections
MicroGenDX NGS detect biofilm bacteria that culture can not.
Many chronic conditions including ENT infections involve biofilms. Biofilms create a barrier, reducing the effectiveness of antibiotics and immune response.
Traditional cultures rarely detect biofilm bacteria and do not identify the full polymicrobial community.
MicroGenDX test identifies biofilm-forming bacteria and fungi via NGS and detects resistance genes via PCR, guiding targeted antibiotic therapy to achieve biofilm embedded pathogens eradication.
Interpreting our lab report
Each test includes a detailed laboratory report, providing clinicians with clear, structured data for fast decision-making.
These reports include:
• List of bacteria and fungi detected in the sample(s)
• Identified antibiotic resistance genes
• Antimicrobials to consideration for treatment



WoundKEY-Test
Wound Infection Test
Wound Swab, Tissue, Fluid
NGS DNA + qPCR Diagnostics for accurate microbial identification
17 antibiotic resistance genes & list of antimicrobials for consideration
3-5 Business days (excludes transit)


NailKEY (MDXNAL)
Nail Infection Test
Nail Clippings
NGS DNA + qPCR Diagnostics for accurate microbial identification
17 antibiotic resistance genes & list of antimicrobials for consideration
3-5 Business days (excludes transit)
Publications Demonstrating the impact of NGS in Wound care
Accelerated healing with targeted therapy guided by MicroGenDX test
| Standard of Care Group | Group 1 | Group 2 |
| Traditional Culture with Systemic Antibiotics | Molecular Diagnostics with Systemic Antibiotics | Molecular Diagnostics with Customized Topical Antibiotics |
48.5%patients healed | 62.4%patients healed | 90.4%patients healed |
NGS yields significantly better outcomes than culture in chronic-wound care. In a level-A retrospective study of ~1,400 cases, MicroGenDX qPCR + NGS–guided systemic antibiotics healed 62.4 % of wounds versus 48.5 % with culture, while topical therapies directed by the same NGS report achieved 90.4 % healing. Time-to-closure was reduced by 26 % and 45.9 %, respectively (P < 0.001).
Culture would have resolved fewer than half the wounds, whereas MicroGenDX molecular guidance healed nearly two-thirds with systemic therapy and nine-tenths with personalised topicals—underscoring its ability to target treatment and accelerate closure.

Chronic wounds: MicroGenDX qPCR+NGS test delivers a more comprehensive polymicrobial profile than culture.
In a prospective Level II study of 30 surgical DFU cases, both methods detected pathogens in 29 samples. NGS revealed a more pathogens than traditional culture. It frequently identified anaerobes such as Finegoldia magna (44.8 %) and Anaerococcus vaginalis (24.1 %), while culture was dominated by Staphylococcus aureus (58.2 %) and missed many fastidious species.
Because culture under-reported key pathogens and resistance genes, its antibiotic guidance was less reliable than NGS. Integrating NGS into DFU management offers comprehensive, species-level data to optimise therapy and enhance limb-salvage outcomes.

NGS delivers more accurate onychomycosis pathogen profile than culture
MicroGenDX NGS testing of 8,816 suspected onychomycosis nails (plus 20 healthy controls) found that ≈ 50 %harboured both fungal + bacterial DNA, 34 % bacteria only, 16 % no detectable organisms, and < 1 % fungi alone. When fungi were present, Trichophyton rubrum dominated but appeared in only 40 % of positives; nondermatophyte fungi Candida, Aspergillus, and Fusarium made up 11 %. None of these fungi appeared in controls, which were mostly benign staphylococci.
The authors emphasize that the MicroGenDX test uncovers fastidious or slow-growing species that culture misses, detecting Candida parapsilosis/ albicans, Aspergillus versicolor/ niger, Fusarium spp., and saprophytes like Pithomyces chartarum, Epicoccum nigrum, and Alternaria alternata. It provides a rapid, quantitative species-level report, giving clinicians a fuller, more actionable pathogen list.

Clinical cases where MicroGenDX test helped achieve treatment success
NGS finds fungus that culture missed to resolve life-threatening wound
A young woman with type I diabetes and a failed kidney transplant was hospitalized twice in six months with unexplained sepsis. She had an open transplant wound and a heel pressure sore, both improving with better care. She later developed sudden left leg pain, swelling, and rapidly spreading erythema. Fasciotomies and drainage revealed only clear, odourless fluid; cultures showed no growth. Despite initial wound improvement, the leg developed necrotic tissue within a week. Fluid sent to MicroGenDX for DNA analysis identified Rhizopus, a cause of mucormycosis (50% mortality). Early detection enabled prompt action by the transplant team.

15-year non-healing leg ulcer closed using MicroGenDX results
A 70-year-old patient had a venous leg ulcer persisting for 15 years, unresponsive to treatments guided by multiple cultures. MicroGenDX testing identified Pluralibacter gergoviae (82%) as the dominant species, along with S. epidermidis (6%), E. coli (5%), and A. vaginalis (2%). No fungi were detected. After targeted compound topical therapy, the wound closed within six weeks and remained closed during follow-up. P. gergoviae, an opportunistic pathogen resistant to antibiotics, has been isolated from clinical and environmental sources. The patient, a former restaurant owner, had prolonged exposure to refrigerated packaged foods.
Inquire about our test
Send us a message using the form below, or email us at info@microgendx.eu
